by John Readey, Senior Architect at The HDF Group
Introduction
HSDS (Highly Scalable Data Service) is a REST-based service for reading and writing HDF data. Initially developed as a NASA Access 2015 project, the HDF Group has continued to invest in the project, and as we’ll see, the latest version has a bevy of new and interesting features.
If you are new to HSDS, it might be useful to review the materials here. Basically though, HSDS endeavors to implement the HDF data model and API in a way that’s suitable for both cloud and on-prem deployments. Here are a few key differentiators between HSDS and the HDF5 library:
- HSDS is a service, i.e. all interactions with it are via REST API vs a library call. As a REST API, the client can be anywhere on the internet which can be useful when you need to provide access to users spread around the globe.
- HSDS (as the name implies) is scalable. The service can take advantage of any number of cores or run on a cluster of machines for even more processing and IO throughput.
- The native storage format is sharded, i.e. each group or dataset is stored as a separate entity (either a POSIX file or object storage object depending on how the deployment is set up). Furthermore, each chunk of each dataset is stored as a separate entity. This organization works well with the cloud (using object storage) and enables features such as multiple writer/multiple reader that aren’t yet supported by the HDF5 library.
You might think: “This is all well and good, but what about my GB’s (or TB’s or PB’s) of HDF5 files and all my applications that use the HDF5 library?”
Regarding existing HDF5 repositories, we provide a utility that can convert HDF5 files to the HSDS format (hsload) as well as a utility that can convert back from HSDS format to a regular HDF5 file (hsget). Any file is “round-tripable” meaning after uploaded and downloading you’ll get an HDF5 file that is logically equivalent to the original (though not binarily identical).
If you have a file collection which you don’t need to modify, there’s an even easier solution: The hsload utility supports a “--link” option which rather than copying all the chunk data just stores pointers into the original file for each chunk. Performance with the link option is comparable to using the native format.
Moving on to application portability we offer two solutions. For Python applications that use the h5py package, there’s the h5pyd package which offers the same api as h5py (with some PyTables-like extensions). To use, you’ll just need to swap out “import h5pyd” with “import h5pyd as h5py” but no other changes should be necessary.
Second, for C or Fortran applications, there’s a plugin for the HDF5 library—the REST VOL. When the REST VOL plugin is installed, applications compiled with the library can use either the native library functionality or have API calls translated to REST requests (based on code to enable the VOL or the presence of an environment variable).
Both h5pyd and the REST VOL support the goal of applications to easily move between “HDF Classic” and HSDS functionality.
Now on to new features in the HSDS 0.6 release…

POSIX storage
Before this release HSDS required an object storage system be used (say S3 on AWS or OpenIO for on-prem installations). While object storage systems have some interesting features (e.g. replication of each object over two or more storage nodes), many users with on-prem systems have existing POSIX storage systems they would like to leverage. POSIX storage is now supported in HSDS and can be as simple as a disk on the server machine or as an elaborate as a Lustre file system cluster. There’s no difference in how clients interact with the server when POSIX storage is used. The only restriction is that Kubernetes deployments are not currently supported (Docker must be used).
Microsoft Azure Support
Posix storage isn’t the only alternative storage system supported in the 0.6 release. To natively integrate with Microsoft Azure we added support for Azure Blob Storage. Azure Blob Storage is an object storage system similar to AWS S3 (but with an entirely different API) and fills a role similar to S3 on AWS: providing a scalable, cost-effective, storage solution.
Also added to support Azure, HSDS authentication can now use Azure Active Directory. Prior to the 0.6 release usernames and passwords were managed by HSDS and HTTP Basic Auth was used for authentication. Azure Active Directory provides a more secure identify provider/authentication scheme (based on OAuth), and reduces the administrative burden of managing user accounts.
And finally, HSDS can be deployed using AKS—Azure Kubernetes Service. This is a managed form of Kubernetes where administrators are no longer burdened with installing (and keep up to date) the Kubernetes Master node.
We’ll be doing a deep dive into the details of HSDS on Azure in a future blog post.
OpenID Authentication
OpenID is an authentication protocol that uses a 3rd party (e.g. Google ID) to perform the authentication. As with Azure Active Directory, when OpenID authentication is setup, the client obtains a token from the authentication server and then passes the token in the HTTP request authentication header to HSDS. HSDS can then securely decode the token and determine the id of the sender. You can find out more about OpenID Authentication here: …
DC/OS Support
DC/OS (https://dcos.io/) is yet another container management platform that derives from Apache Mesos. It can be used for both on-prem and cloud deployments.
DC/OS support was provided thanks to a community contribution. This might be a good time to mention that The HDF Group welcomes contributions to HSDS and supported projects (and to the library itself for that matter). HSDS and h5pyd have been on GitHub since the projects started, and we’ve found GitHub to be an excellent platform for different groups to collaborate. If you have an interesting feature you’d like to add to HSDS, please start by creating an issue in the github repo.
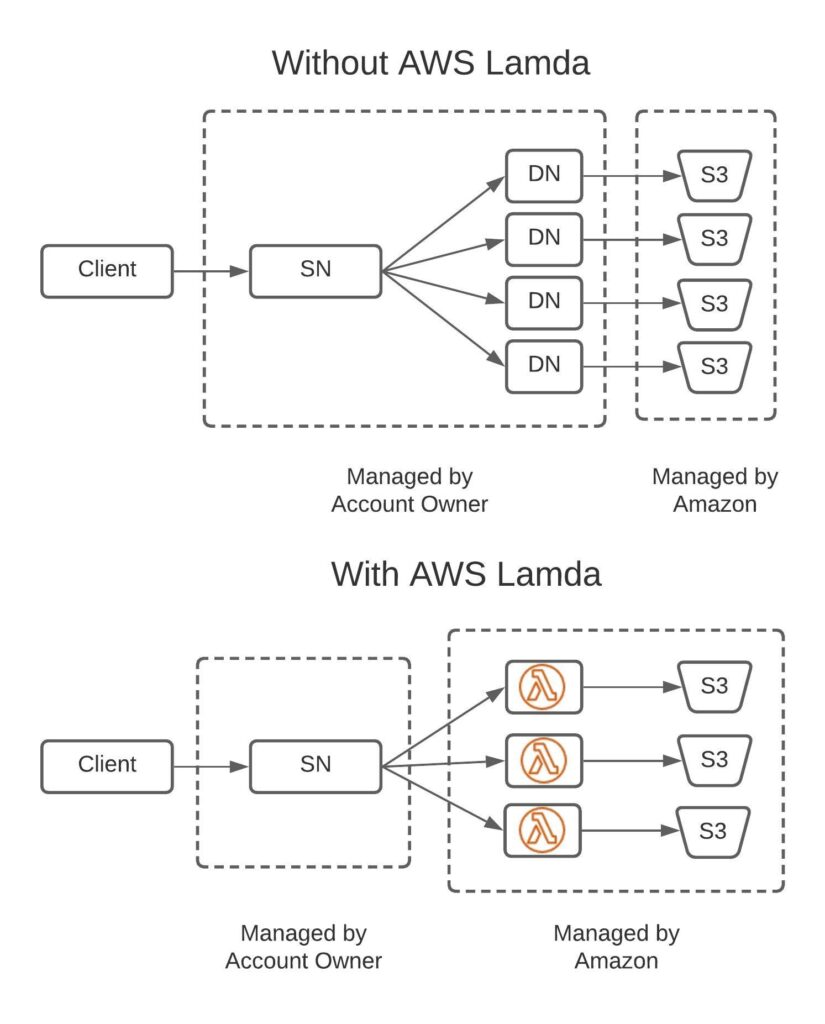
AWS Lambda Support
AWS Lambda is a service that enables user defined functions to be run without a server. Of course the function is run on a computer, but when using Lambda it’s Amazon that manages a fleet of servers for you. This fleet has sufficient compacity for almost any scenario a customer may cook up. Each time a Lambda function is invoked, Amazon will charge your account for the amount of time it took to run (currently: $0.00001667/GB-sec). Since you only pay for the time your function is running this is generally more cost effective than setting up your own cluster. Also it removes the devops burden with managing your own cluster.
For HSDS we haven’t completely replaced the existing software using Lambda functions (for one thing it wouldn’t be possible to support multi-client write operations using AWS Lambda), but Lambda is used as a sort of “turbo boost” for certain operations.
Consider a client which is doing a hyperslab selection on a dataset. Depending on the chunk layout of the dataset and the range of the hyperslab, this selection may touch one, a dozen, or a thousand chunks (or anywhere in between). If HSDS is running with say 4 nodes, the dozen chunk case is handled very efficiently—each node will be assigned a portion of the chunks need to service the request. The nodes in turn fetch the chunk, segment and return the desired data. This enables a nice degree of parallelism since the work of reading the chunks is spread out across multiple processes (when running on Docker) or potentially multiple machines (when running on Kubernetes or DC/OS).
Now consider the thousand chunk selection case. If HSDS is running on 4 nodes, each node would have about 250 chunks to deal with, which would take significant time to process. Potentially, you can scale up HSDS to run on 100’s of nodes to speed things up but it would consume considerable resources running such a large deployments. If this type of selection is fairly infrequent, utilization of the server(s) will be low, which is not cost-effective. Alternative you can scale up the HSDS deployment as needed, but that action can’t happen fast enough to service a particular request.
This is where AWS Lambda is a useful addition—when Lambda is configured for the HSDS deployment, HSDS can run up to 1000 chunk read functions in parallel on Lambda. Each AWS region will have a server pool large enough to handle such spike loads immediately, so in effect it’s like having a 1000 server cluster that pops into existence just as you need it.
Lambda is not always faster than just having HSDS do the chunk reads. Lambda functions have a startup latency that may out weigh the benefits of increased parallelism. Results will depend on how your datasets are structured and the type of selections used. The best advice will be to try HSDS configured with Lambda vs without and see which works better for you.
Of course this feature is only supported for AWS deployments. Azure has a similar feature “Azure functions” that we’d like to support as well, but that didn’t make it into this release.
BLOSC Compression
The HDF5 library supports only Deflate and SZIP compression “out of the box”. You can add custom compression filters using the compression plugin support, but this can be a bit of a hassle to setup. Also, if you used a custom compression filter, and then share the resulting file, you’ll need to make sure each reader has the filter installed as well.
With HSDS things are a bit simpler, since if the server support a certain compression filter, clients don’t need any special software to use it – compression happens on the server side.
HSDS has supported the deflate compressor (the same as with the HDF5 library), but with the 0.6 release we added support for BLOSC compression as well. BLOSC is the brainchild of our HDF friend and sometimes collaborator: Francesc Alted (see this blog post). BLOSC is very efficient and has been available for use with the HDF5 for many years (notably the PyTables package comes with BLOSC).
A nice aspect of BLOSC is that it’s a “metacompressor”. That is BLOSC includes a number of different compression formats: lzf, blosclz, snappy, lz4, and zstd, so it’s like having five compressors for the price of one! BLOSC also supports mutithreading, so compression/decompression can take advantage of available cores and hyper threading.
Utilizing BLOSC in the HSDS 0.6 release is quite simple. Nothing special needs to be done to read datasets compressed with BLOSC. To create a file using one of the BLOSC compressors in Python, just use create_dataset with the compressor option set to the desired compressor. With the REST VOL, use the appropriate H5Z_FiLTER_ID - see.
Role Based Access Control (RBAC)
As explained in To Serve and Protect: Web Security for HDF5, HSDS uses Access Control Lists (ACLs) to authorize certain actions (read,w rite, delete, etc.) for specific users. The set of ACLs for a particular file (domain in HSDS-speak) is stored along with the domain metadata. The hsacl tool can be used to view or modify the domains ACLs (the REST API can also be used).
This setup works well enough, but can be awkward if their a many users to manage—e.g. suppose there are 20 users in the engineering department who should all have access to a set of domains. Rather than setting up 20 ACLs per domain (and updating these as the members of the organization change), you can define the set of users in HSDS and then just create an ACL that defines that set’s permissions. If the group’s (group here in the sense of a group of users, not an HDF5 group) name is “eng_dept”, then you can just do for example: “`hsacl /some_path/some_domain.h5 +r g:eng_dept`“. This will give everyone in the eng_dept group permissions to read the some_domain.h5 domain. The “g:” prefix is to disambiguate group ACLs from user level ACLs.
Domain Checksums
For regular files, a checksum is a useful way to determine if the contents of the file have changed or not. On Linux you can use the “`cksum [file]`” to get the checksum for any particular file.
With HSDS doing an equivalent operation on a domain is a bit more challenging. Firstly, as an HSDS user, you may not have direct access to the storage system being used. Secondly, a single HSDS domain may contain millions of files (for POSIX deployments) or objects (for object storage deployment).
To provide a checksum feature, for the 0.6 release, we added a server features that computes a per domain checksum anytime the domain contents are updated. The checksum computation runs asynchronously, so it may take some seconds for the checksum to update when the domain contents are changed.
You can view the checksum via run the hsinfo command with the domain path. Example:
`$ hsinfo /home/john/terra2/TERRA_BF_L1B_O10307_20011125025026_F000_V001.h5 domain: /home/john/terra2/TERRA_BF_L1B_O10307_20011125025026_F000_V001.h5 owner: john id: g-84a0ff3c-e25dcfbd-0c28-77b46a-d54735 last modified: 2020-07-09 22:26:05 last scan: 2020-07-09 22:26:33 md5 sum: f8c4f6efba9c517b35c2381a72bae8fe total_size: 17133812060 allocated_bytes: 11256 metadata_bytes: 1461174 linked_bytes: 17132339378 num objects: 929 num chunks: 60 linked chunks: 269741`
From the output we can see that this domain has 929 objects (i.e. datasets or groups), 60 chunks, and 269,741 linked chunks (chunks that are located in a HDF5 file). The md5 sum is a sum of sums over the more than 270 thousand objects in the domain. If any dataset values, attribute, or link is modified, the checksum will be different.
Also notice that the domain was last modified on July 9th, 2020 at 10:26:05PM and the “last scan” time (the time in which the checksum was recomputed) was at 10:26:35, so in this case there was a 30 second lag between the time the domain was modified and the new checksum was available.
Conclusions
This concludes our tour of some of the new features in the HSDS 0.6 release. (For the complete list of new features see: https://github.com/HDFGroup/hsds/issues/47) HSDS is free and open source, so if your curiosity is piqued, please go to https://github.com/HDFGroup/hsds for instructions on setting up your own HSDS server. We’d appreciate any feedback you might have on this release!